Command line tools for quality control
Figure 1
The structure of a typical command line
entry.
Figure 2
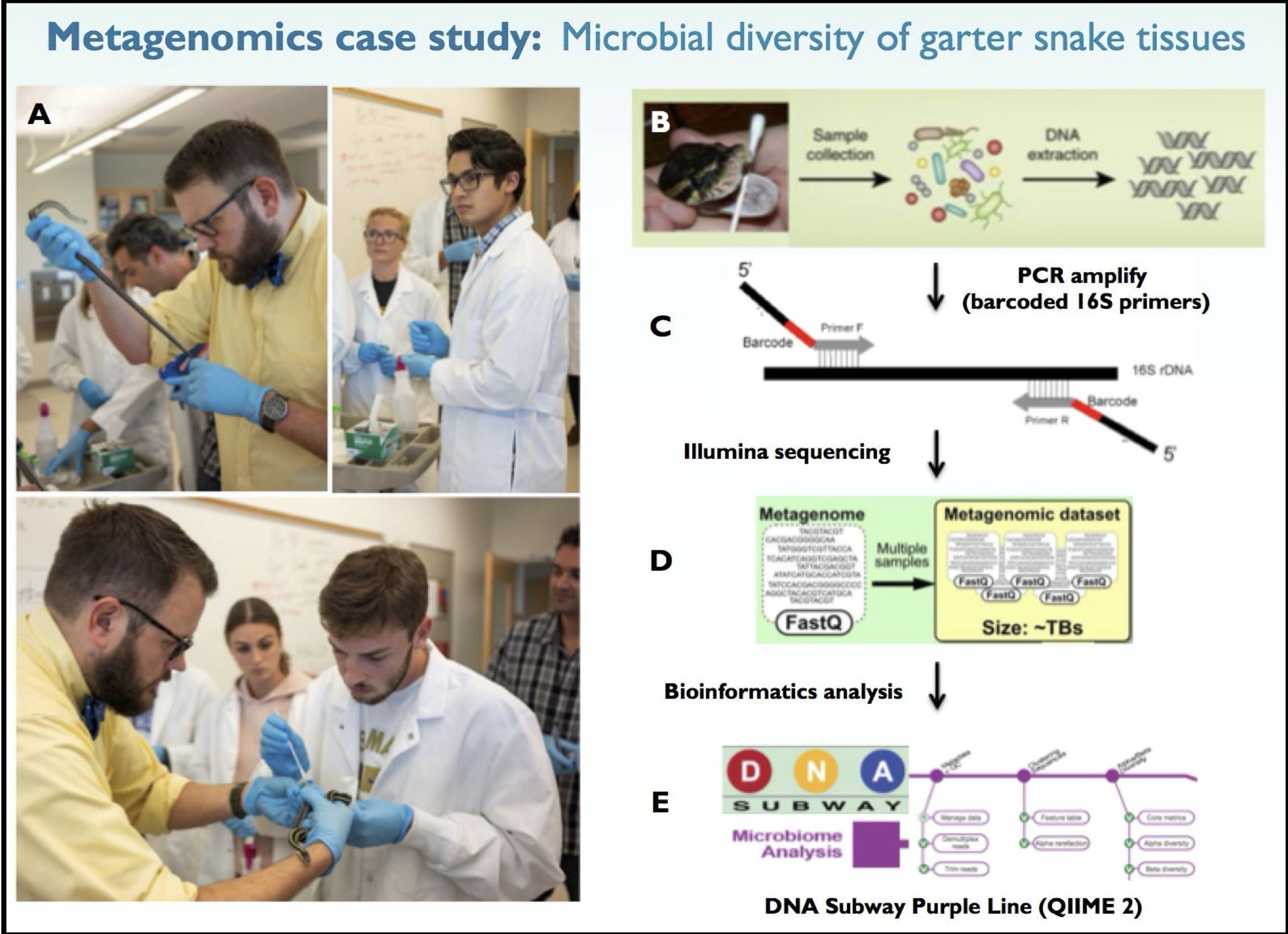
Figure 1. Overview of the in class metagenomics
project. Using a saline swabbing technique, microbial samples were
collected from garter snake tissues in class (A). Swabs were placed in
sterile tubes to release collected microbes and DNA was extracted for
downstream analysis (B). Barcoded primers were used to PCR amplify the
microbial 16S ribosomal DNA repeat genes for each sample followed by
Illumina sequencing of PCR amplicons (C-D). The DNA Subway Purple Line
web-based software can be used to analyze FASTQ data files generated
from Illumina sequencing to reveal the microbial population of our swabs
(E). Garter snakes were provided by Dr. Rocky Parker in the JMU
Department of Biology (A; yellow shirt).
